There are essentially three graph implementations, listed below
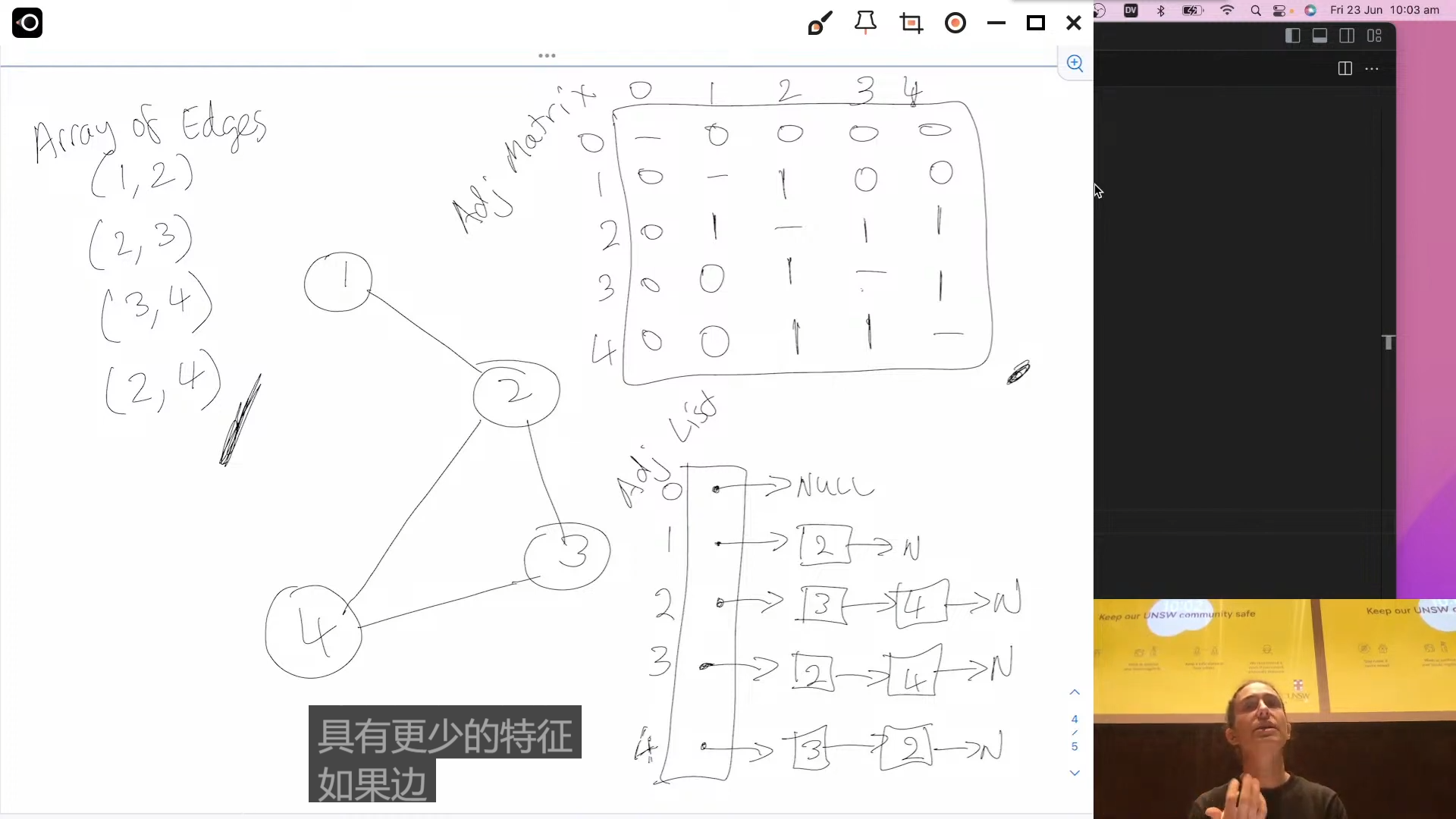
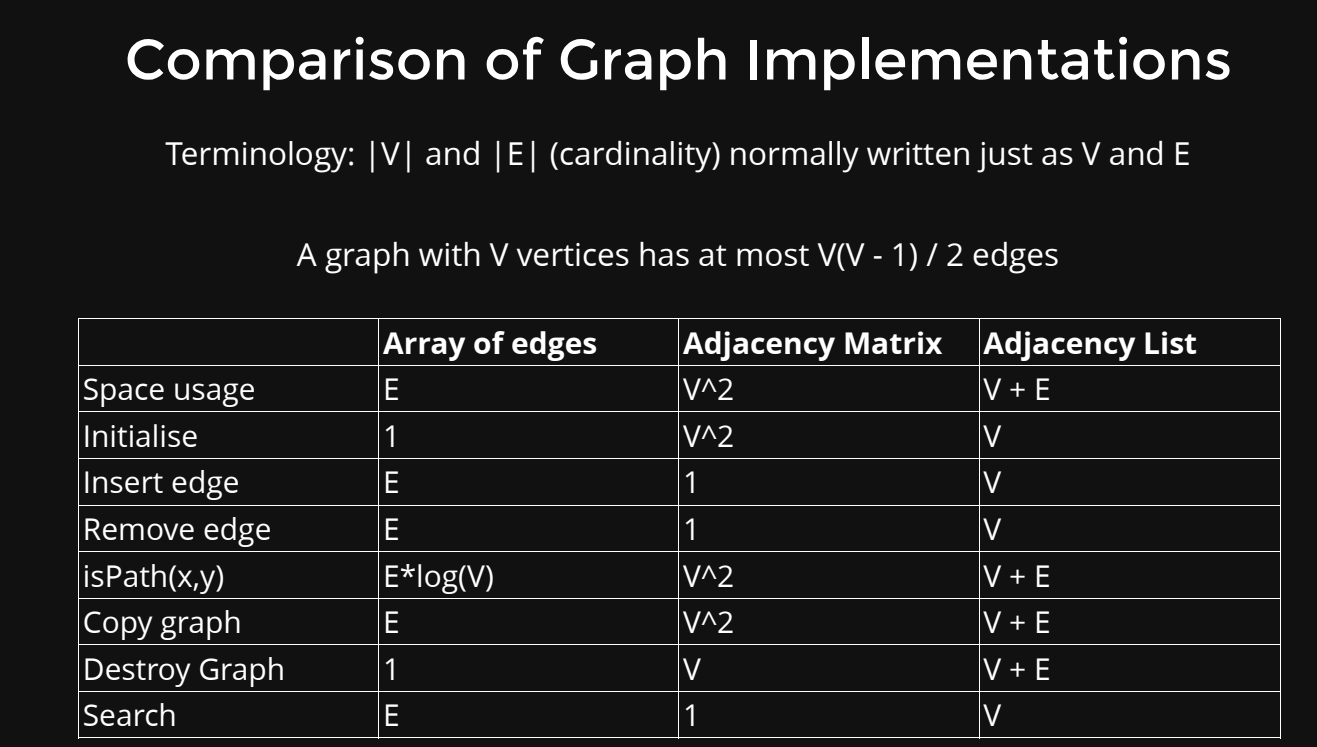
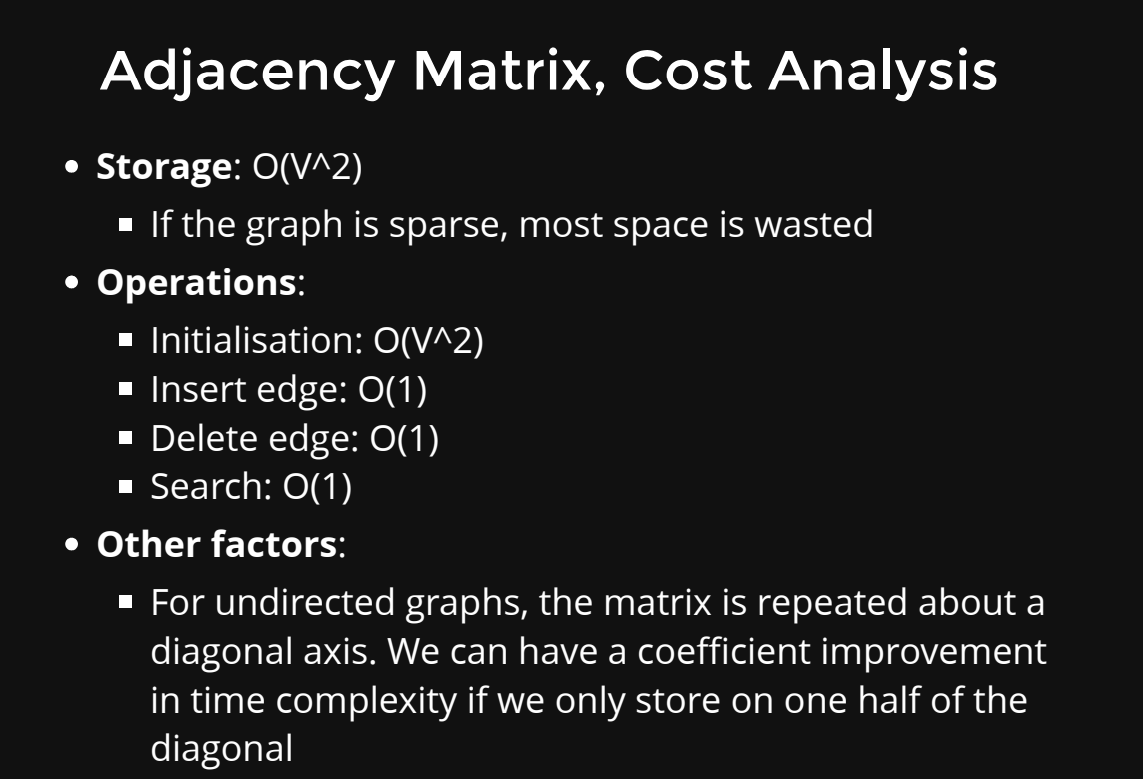
- Adjacency Matrix: An adjacency matrix is a two-dimensional array where each cell (i, j) represents whether there is an edge between node i and node j. If the graph is weighted, the cell can store the weight of the edge. The value in the cell can be a boolean (for unweighted graphs) or a number (for weighted graphs).
Advantages:
- Constant-time O(1) access to check if an edge exists between two nodes.
- Efficient for dense graphs (graphs with many edges).
- Suitable for graphs with a fixed number of nodes.
Disadvantages:
- Consumes more space for sparse graphs (graphs with few edges).
- Adding or removing nodes requires resizing the matrix, which can be costly.
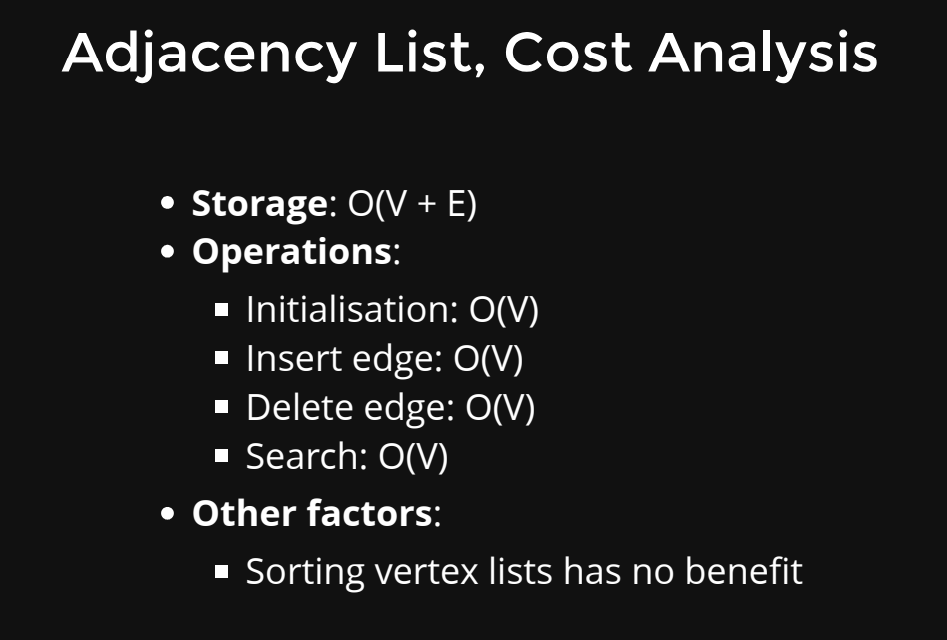
- Adjacency List: An adjacency list is a data structure where each node is associated with a list (or array) of its neighboring nodes. For a weighted graph, each element in the list can store the weight of the edge.
Advantages:
- Memory-efficient for sparse graphs.
- Efficient for graph algorithms that involve traversing neighbors.
- Adding or removing nodes/edges is more efficient than with an adjacency matrix.
Disadvantages:
- Slower edge existence check (requires traversing the adjacency list).
- May consume more memory for dense graphs.
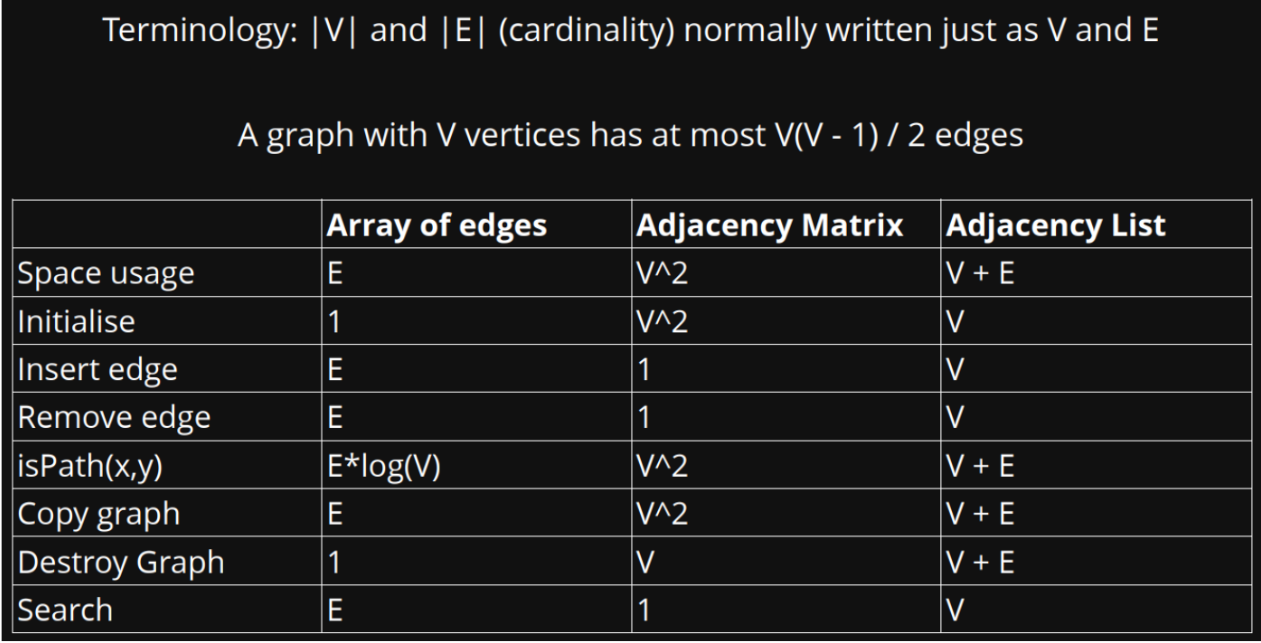
- Array of Edges (Edge List): An array of edges represents the graph as an array where each element represents an edge. Each edge is defined by its two endpoints (nodes) and possibly a weight.
Advantages:
- Minimal memory overhead.
- Simple structure, suitable for basic graph representations.
- Good for storing just the essential information about the graph.
Disadvantages:
- Edge existence check can be inefficient if the array is not sorted or indexed.
- Traversing neighbors may require additional data structures.